Sobre
A idéia principal aqui é coletar dados ECG com a intenção de tentar prever crises epiléticas. Mais sobre o projeto em Dispositivo para prever crises epiléticas.
O estudo está “on going”, vou atualizando essa nota com updates.
Por agora, apenas testes com a controladora e sensor. A próxima etapa é saber como lidar com os dados. Já vi uma possibilidade usando talvez neurokit.
Depois, possivelmente podemos usar um MQTT para receber os dados do dispositivo e processa-los. Nesse ponto, parece ser possível usar técnicas descritas nesses artigos:
- An approach to detect and predict epileptic seizures with high accuracy using CNN and single-lead-ECG signal.
- Wearable Epileptic Seizure Prediction System with Machine-Learning-Based Anomaly Detection of Heart Rate Variability
- ECG-Based Semi-Supervised Anomaly Detection for Early Detection and Monitoring of Epileptic Seizures
Implementação
Usei uma ESP32 como controladora e Placa AD8232 para coleta do sinal analógico dos Eletrodos.
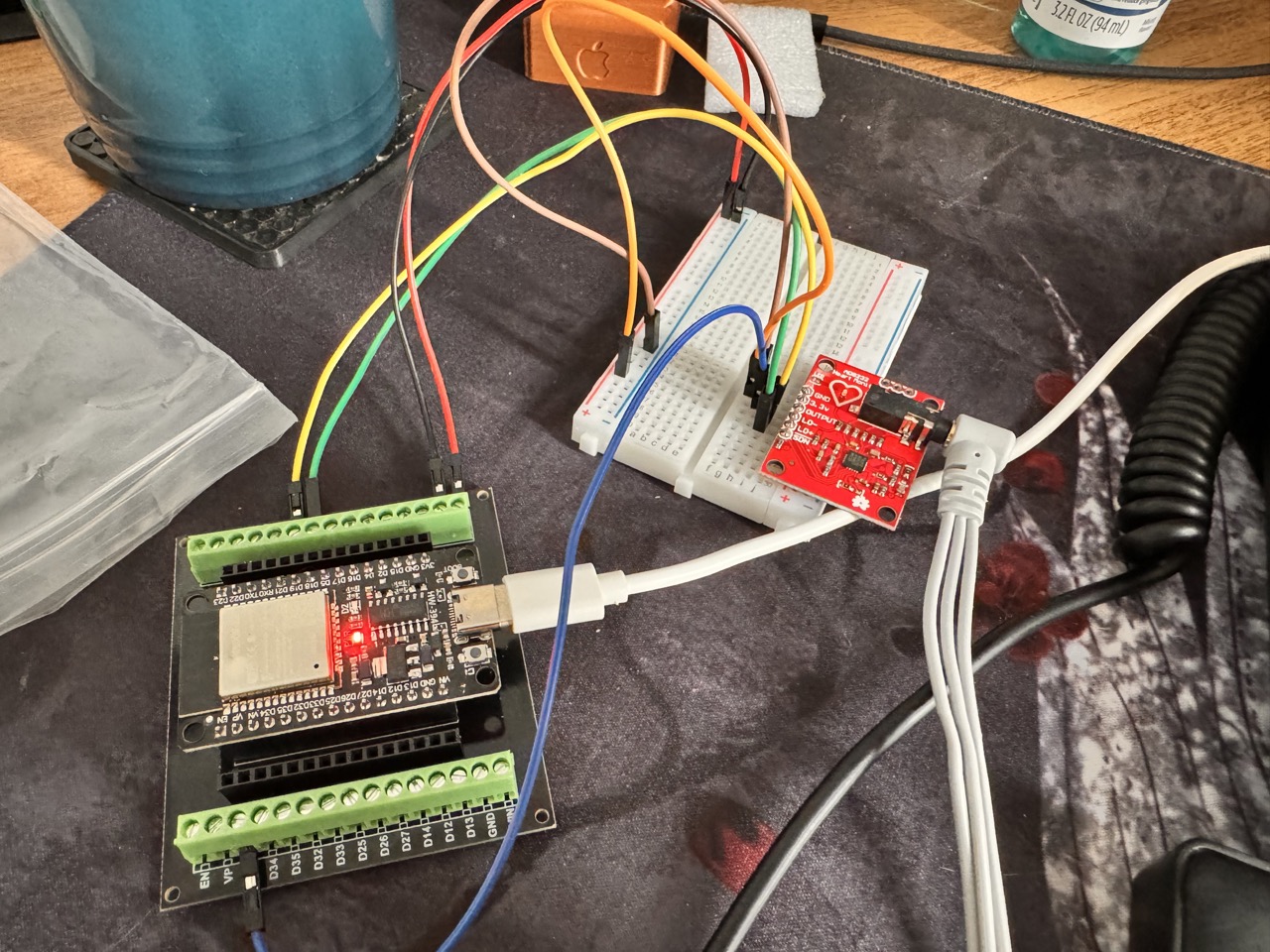
Exemplo de código com ESP32
Fonte: 1 (com adaptação do código arduino para ESP32 pelo modelo OpenAI O1)
Minha conexão dos pinos entre as placas:
Lo- (amarelo): D19 Lo+ (verde): D18 Output (analog): 39 - Usando GPIO39 (Porta VN na ESP32)
/*
* ECG Data Collection using the AD8232 module with ESP32.
* This code collects the ECG signal and applies a digital notch filter to eliminate
* 60Hz power line noise.
* The built-in LED lights up when the AD8232 module detects a loose electrode.
*
* Notch filter based on:
* CHOY, T. T. C.; LEUNG, P. M. Real-time microprocessor-based
* 50 Hz notch filter for ECG. Journal of Biomedical Engineering,
* v. 10, n. 3, p. 285-288, 1988.
*
* Filter options vary with the sampling frequency (Fs):
* For Fs = 120Hz, use "N_POINTS 2" and "N 1"
* For Fs = 240Hz, use "N_POINTS 3" and "N 2"
* For Fs = 360Hz, use "N_POINTS 4" and "N 3"
* "a" should be between 0 and 1:
* - "a" close to 0 distorts the ECG signal more but is less susceptible to variations
* in noise and sampling frequencies.
* - "a" close to 1 distorts the ECG signal less but is more susceptible to variations
* in noise and sampling frequencies.
*
* Created by: Erick Dario León Bueno de Camargo, 2023
* Adapted by: [Your Name], 2023
*/
// Desired sampling frequency in Hz:
#define SAMPLE_FREQUENCY 120
// Constants for the notch filter
#define N_POINTS 2
#define N 1
#define a 0.05
// Pin definitions based on your hardware setup
#define ANALOG_PIN 39 // Use GPIO39 (Port VN in) (ADC1_CH0) for analog input
#define LO_MINUS_PIN 19 // GPIO19 for "Lo-"
#define LO_PLUS_PIN 18 // GPIO18 for "Lo+"
#define LED_PIN 2 // Built-in LED (adjust if necessary)
unsigned long sample_interval_us = 1000000UL / SAMPLE_FREQUENCY;
int X[N_POINTS]; // Array for current measurements
int Y[N_POINTS]; // Array for filtered measurements
unsigned long current_time, previous_time;
int counter = 0;
void setup() {
Serial.begin(115200);
pinMode(LO_MINUS_PIN, INPUT);
pinMode(LO_PLUS_PIN, INPUT);
pinMode(LED_PIN, OUTPUT);
previous_time = micros();
}
void loop() {
current_time = micros();
// Check for loose electrodes
if ((digitalRead(LO_MINUS_PIN) == HIGH) || (digitalRead(LO_PLUS_PIN) == HIGH)) {
digitalWrite(LED_PIN, HIGH);
} else {
digitalWrite(LED_PIN, LOW);
// Sampling at precise intervals
if (current_time - previous_time >= sample_interval_us) {
previous_time += sample_interval_us; // Maintain consistent sampling rate
// Read analog value from the ECG module
X[counter] = analogRead(ANALOG_PIN);
int previous_index = (counter + N_POINTS - N) % N_POINTS;
// Apply notch filter
Y[counter] = ((X[counter] + X[previous_index]) / 2) +
a * (((X[counter] + X[previous_index]) / 2) - Y[previous_index]);
// Output the filtered signal
Serial.println(Y[counter]);
// Update counter
counter = (counter + 1) % N_POINTS;
}
}
}Exemplo de dados coletados
Python para coleta dos dados da porta serial usando a lib pyserial.
import serial
import time
# Adjust the serial port and baud rate to match your setup
# ser = serial.Serial('COM3', 115200) # For Windows, e.g., 'COM3'
# ser = serial.Serial('/dev/ttyUSB0', 115200) # For Linux
ser = serial.Serial('/dev/tty.usbserial-112220', 115200) # For macOS
# Create or open a file to write the data
with open('./ecg_data.csv', 'w') as file:
# Write header if needed
file.write('timestamp,data\n')
# Collect data for a specified duration or until stopped
try:
while True:
# Read a line from the serial port
line = ser.readline().decode('utf-8').strip()
# Get the current timestamp
timestamp = time.time()
# Write timestamp and data to the file
file.write(f'{timestamp},{line}\n')
# Print to the console for monitoring (optional)
print(f'{timestamp},{line}')
except KeyboardInterrupt:
print("Data collection stopped by user.")
# Close the serial port
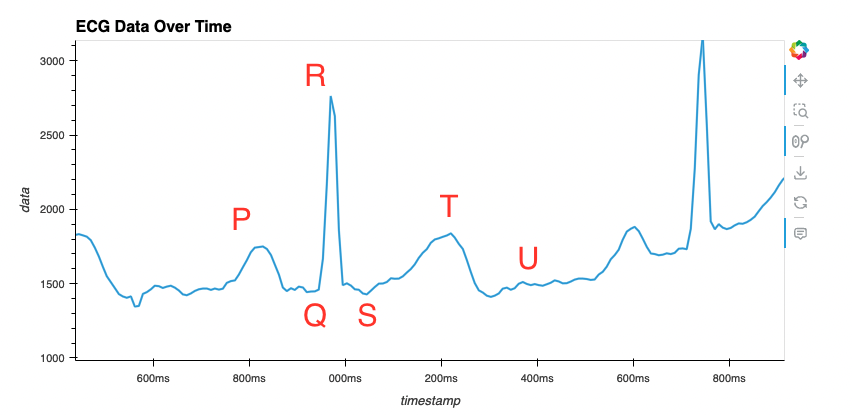
ser.close()Sample rate de 120hz Dados: ecg_data_120.csv
Sample rate de 360hz Dados: ecg_data_360.csv
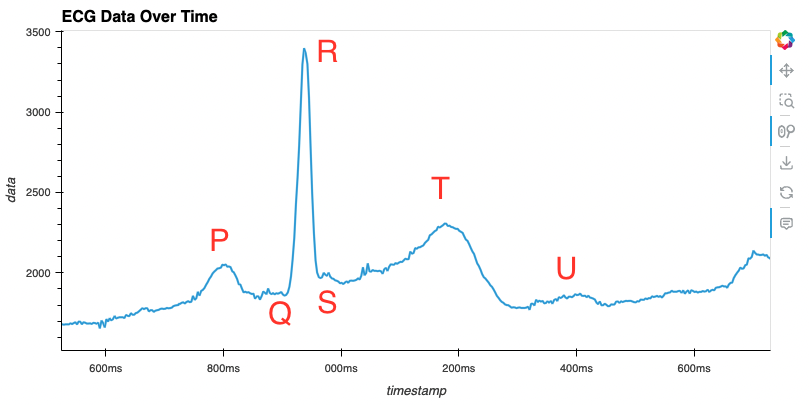
Código pandas com hvplot usado para “plotar” os dados:
import pandas as pd
import hvplot.pandas # hvPlot extension
# Load the ECG data
file_path = 'ecg_data.csv'
ecg_data = pd.read_csv(file_path)
# Convert the timestamp to a more readable format if necessary
# Assuming timestamp is in seconds, convert it to datetime
ecg_data['timestamp'] = pd.to_datetime(ecg_data['timestamp'], unit='s')
# Convert 'data' to numeric, forcing errors to NaN (e.g., "S1774" is invalid)
ecg_data['data'] = pd.to_numeric(ecg_data['data'], errors='coerce')
# Create an interactive plot using hvPlot
ecg_data.hvplot(x='timestamp', y='data', title='ECG Data Over Time', width=800, height=400)Interpretação do ECG
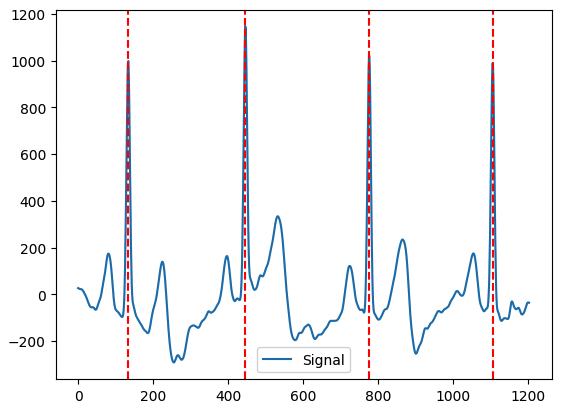
Usando a lib neurokit
import neurokit2 as nk
values = ecg_data['data'].values
cleaned_ecg = nk.ecg_clean(values, sampling_rate=360) # 360hz
_, rpeaks = nk.ecg_peaks(cleaned_ecg, sampling_rate=360)
plot = nk.events_plot(rpeaks['ECG_R_Peaks'], cleaned_ecg)Output: